Technical development
CGI Extravaganza! pulling categories from mediawiki(SNPedia)
- local cgi with Fako & Natasha (feb,15th). From the terminal (local host), this python script pulls categories (default on 10) from SNPedia.
#!/usr/bin/python
import json, urllib2
search = u'Rs1001179' # quotes are string.
url = u'http://www.snpedia.com/api.php?action=query&format=json&redirects=true&generator=categories&titles=' + search
request = urllib2.Request(url)
response = urllib2.urlopen(request)
result = response.read()
f = json.loads(result)
print type(f)
g = json.dumps(f) # dump makes back into a string
print type(g)
for page in f['query']['pages'].itervalues():
print page['title']
-Meaning that rsid number Rs1001179 is found in 10 categories in following results (terminal):
<type 'dict'>
<type 'str'>
Category:On chip Illumina Human 1M
Category:SNPs on chromosome 11
Category:In dbSNP
Category:Is a snp
Category:On chip 23andMe v3
Category:On chip HumanOmni1Quad
Category:On chip 23andMe v2
Category:Has population
Category:On chip 23andMe v1
Category:Has genotype
- Server cgi with Michael (evening of feb,15th), viewed in browser
-first connect to the pzwart3 server in terminal:
sshfs awu@pzwart3.wdka.hro.nl:public_html mnt
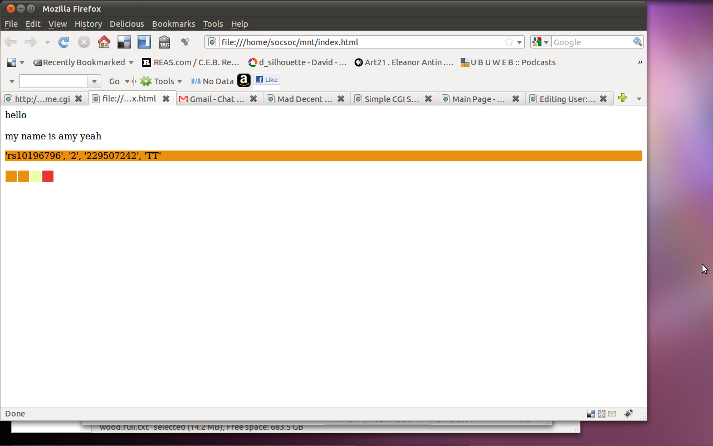
-in index.html i define the parameters:
hello <br /><br />
my name is amy yeah
<p style="background: #ea9010; ">'rs10196796', '2', '229507242', 'TT'</p>
<div style="float: left; width: 20px; height: 20px; background: #ea9010; margin: 1px"></div>
<div style="float: left; width: 20px; height: 20px; background: #ea9010; margin: 1px"></div>
<div style="float: left; width: 20px; height: 20px; background: #eaffaa; margin: 1px"></div>
<div style="float: left; width: 20px; height: 20px; background: #ea3333; margin: 1px"></div>
<!-- http://pzwart3.wdka.hro.nl/~awu/ -->
visualisation:
(note that url is: file:///home/socsoc/mnt/index.html)
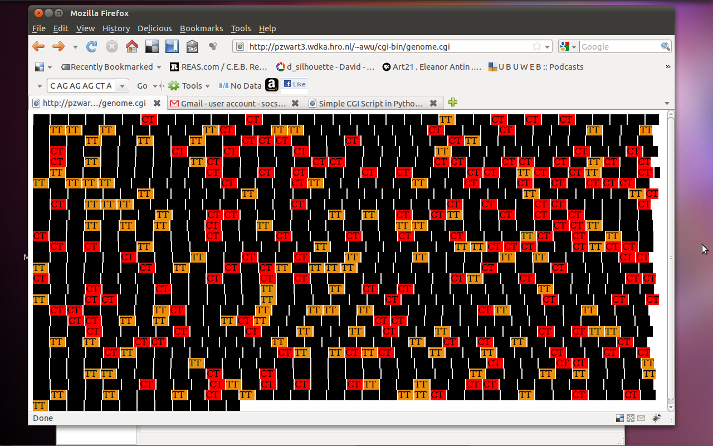
-and use python as a 'motor' or 'conveyor belt' to mass produce the result:
#!/usr/bin/env python
#-*- coding:utf-8 -*-
import cgitb; cgitb.enable()
print "Content-Type: text/html; charset=utf-8" # HTML is following
print
f = open ('wood.txt')
data = []
for line in f:
if not line.startswith ('#'):
d = line.strip().split()
if len(d) == 4:
data.append(d)
colors = {
'TT': '#ea9010',
'CT': '#FF0000'
}
for (rsid, chromosome, position, genotype) in data:
print """<span style="width: 20px; background: """,
print colors.get(genotype, 'black'),
print """; margin: 1px">""",
#print " ",
print genotype,
print "</span>"
print
visualisation:
(note that url is: http://pzwart3.wdka.hro.nl/~awu/cgi-bin/genome.cgi)